6.3 Dimensionality Reduction After Harmony
In a previous chapter, we performed batch correction using Harmony via the addHarmony() function, creating a reducedDims object named “Harmony”. We can assess the effects of Harmony by visualizing the embedding using UMAP or t-SNE and comparing this to the embeddings visualized in the previous sections for iterative LSI.
Repeating the UMAP embedding with the same parameters but for the “Harmony” reducedDims object:
projHeme2 <- addUMAP(
ArchRProj = projHeme2,
reducedDims = "Harmony",
name = "UMAPHarmony",
nNeighbors = 30,
minDist = 0.5,
metric = "cosine"
)## 09:53:44 UMAP embedding parameters a = 0.583 b = 1.334
## 09:53:44 Read 10251 rows and found 30 numeric columns
## 09:53:44 Using Annoy for neighbor search, n_neighbors = 30
## 09:53:44 Building Annoy index with metric = cosine, n_trees = 50
## 0% 10 20 30 40 50 60 70 80 90 100%
## [—-|—-|—-|—-|—-|—-|—-|—-|—-|—-|
## **************************************************|
## 09:53:47 Writing NN index file to temp file /tmp/RtmpXb8qQa/fileefee55b054c6
## 09:53:47 Searching Annoy index using 10 threads, search_k = 3000
## 09:53:48 Annoy recall = 100%
## 09:53:49 Commencing smooth kNN distance calibration using 10 threads
## 09:53:50 Initializing from normalized Laplacian + noise
## 09:53:51 Commencing optimization for 200 epochs, with 472754 positive edges
## 09:54:01 Optimization finished
p3 <- plotEmbedding(ArchRProj = projHeme2, colorBy = "cellColData", name = "Sample", embedding = "UMAPHarmony")## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-efeea6eac67-Date-2020-04-15_Time-09-54-03.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = cellColData
## Plotting Embedding
## 1
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-efeea6eac67-Date-2020-04-15_Time-09-54-03.log
p4 <- plotEmbedding(ArchRProj = projHeme2, colorBy = "cellColData", name = "Clusters", embedding = "UMAPHarmony")## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-efee76ae6a66-Date-2020-04-15_Time-09-54-04.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = cellColData
## Plotting Embedding
## 1
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-efee76ae6a66-Date-2020-04-15_Time-09-54-04.log
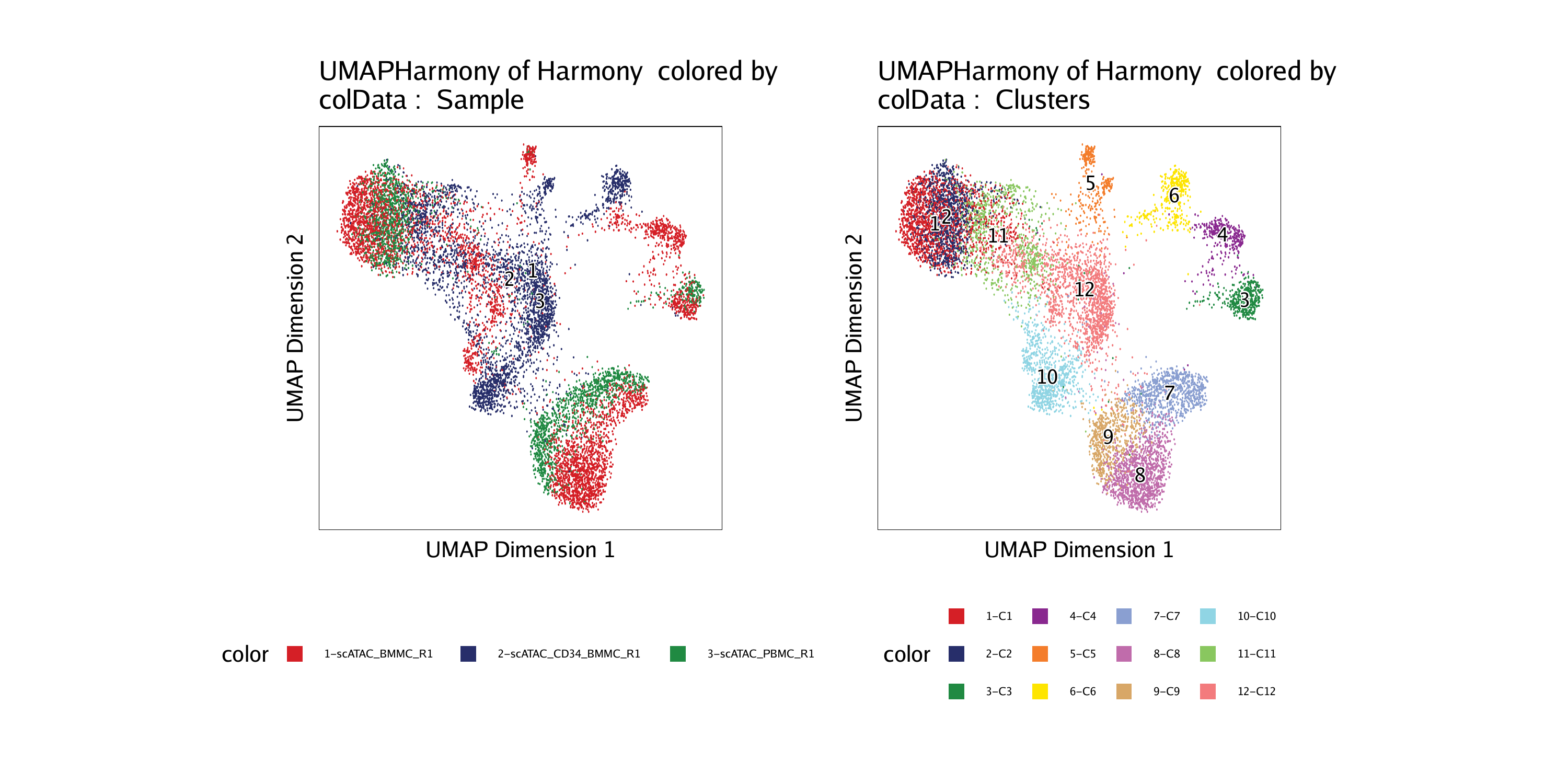
To save an editable vectorized version of this plot, we use plotPDF().
plotPDF(p1,p2,p3,p4, name = "Plot-UMAP2Harmony-Sample-Clusters.pdf", ArchRProj = projHeme2, addDOC = FALSE, width = 5, height = 5)## [1] “plotting ggplot!”
## [1] “plotting ggplot!”
## [1] “plotting ggplot!”
## [1] “plotting ggplot!”
## [1] 0
Download PDF : Plot-UMAP2Harmony-Sample-Clusters.pdf
And the same for t-SNE:
projHeme2 <- addTSNE(
ArchRProj = projHeme2,
reducedDims = "Harmony",
name = "TSNEHarmony",
perplexity = 30
)## Read the 10251 x 30 data matrix successfully!
## OpenMP is working. 9 threads.
## Using no_dims = 2, perplexity = 30.000000, and theta = 0.500000
## Computing input similarities…
## Building tree…
## - point 10000 of 10251
## Done in 12.20 seconds (sparsity = 0.013705)!
## Learning embedding…
## Iteration 50: error is 95.841545 (50 iterations in 9.09 seconds)
## Iteration 100: error is 86.878252 (50 iterations in 10.44 seconds)
## Iteration 150: error is 84.458350 (50 iterations in 6.52 seconds)
## Iteration 200: error is 84.214403 (50 iterations in 6.81 seconds)
## Iteration 250: error is 84.119663 (50 iterations in 6.93 seconds)
## Iteration 300: error is 3.435665 (50 iterations in 6.05 seconds)
## Iteration 350: error is 3.186510 (50 iterations in 5.82 seconds)
## Iteration 400: error is 3.055847 (50 iterations in 5.83 seconds)
## Iteration 450: error is 2.971219 (50 iterations in 5.78 seconds)
## Iteration 500: error is 2.911368 (50 iterations in 5.75 seconds)
## Iteration 550: error is 2.867716 (50 iterations in 5.70 seconds)
## Iteration 600: error is 2.833307 (50 iterations in 5.75 seconds)
## Iteration 650: error is 2.806291 (50 iterations in 5.72 seconds)
## Iteration 700: error is 2.784594 (50 iterations in 5.68 seconds)
## Iteration 750: error is 2.767633 (50 iterations in 5.68 seconds)
## Iteration 800: error is 2.754284 (50 iterations in 5.72 seconds)
## Iteration 850: error is 2.743628 (50 iterations in 5.71 seconds)
## Iteration 900: error is 2.735122 (50 iterations in 5.71 seconds)
## Iteration 950: error is 2.727634 (50 iterations in 5.67 seconds)
## Iteration 1000: error is 2.721500 (50 iterations in 5.69 seconds)
## Fitting performed in 126.05 seconds.
p3 <- plotEmbedding(ArchRProj = projHeme2, colorBy = "cellColData", name = "Sample", embedding = "TSNEHarmony")## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-efee50fc7167-Date-2020-04-15_Time-09-55-42.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = cellColData
## Plotting Embedding
## 1
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-efee50fc7167-Date-2020-04-15_Time-09-55-42.log
p4 <- plotEmbedding(ArchRProj = projHeme2, colorBy = "cellColData", name = "Clusters", embedding = "TSNEHarmony")## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-efee8ad75d2-Date-2020-04-15_Time-09-55-42.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = cellColData
## Plotting Embedding
## 1
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-efee8ad75d2-Date-2020-04-15_Time-09-55-42.log
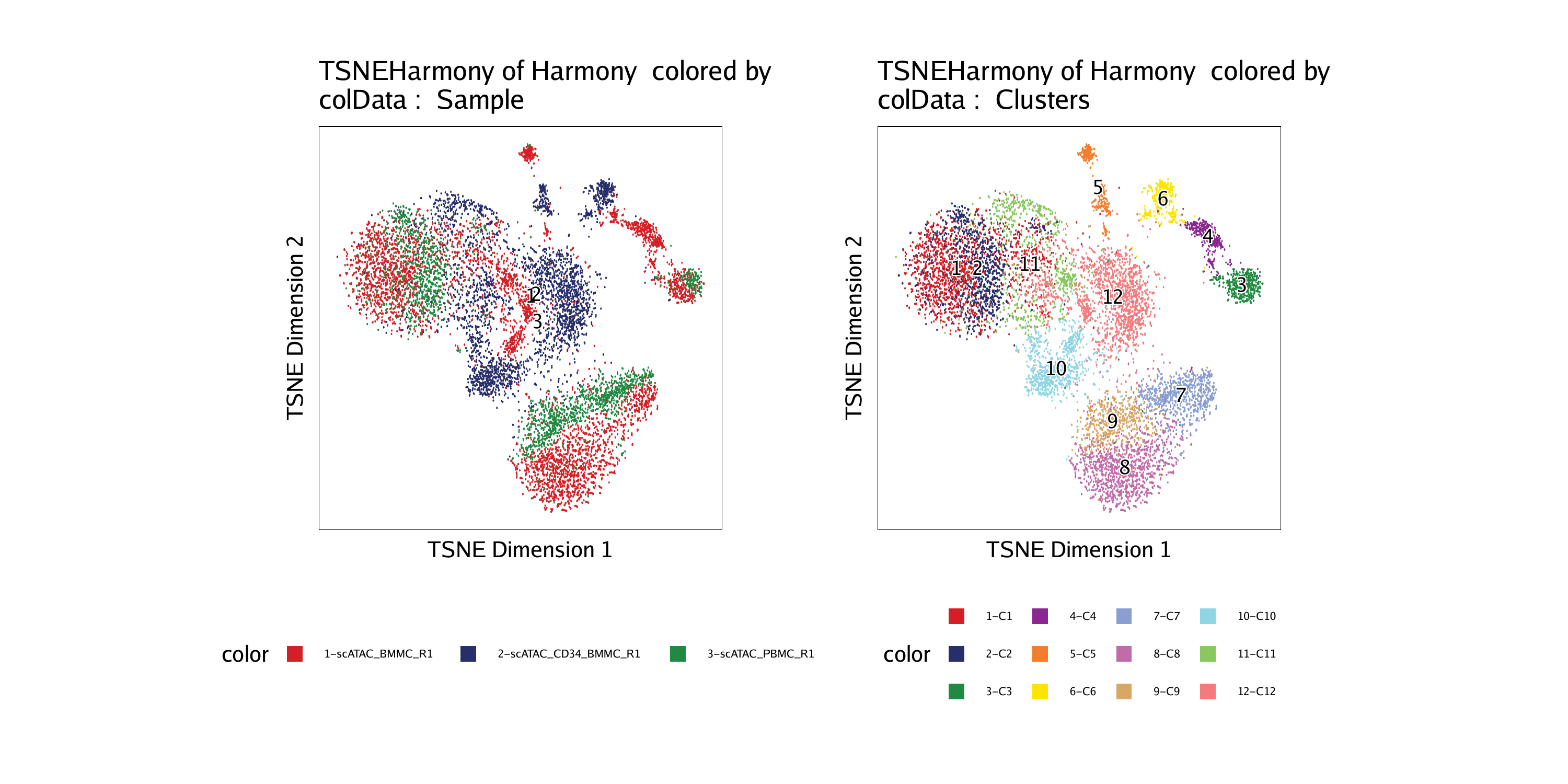
To save an editable vectorized version of this plot, we use plotPDF().
plotPDF(p1,p2,p3,p4, name = "Plot-TSNE2Harmony-Sample-Clusters.pdf", ArchRProj = projHeme2, addDOC = FALSE, width = 5, height = 5)## [1] “plotting ggplot!”
## [1] “plotting ggplot!”
## [1] “plotting ggplot!”
## [1] “plotting ggplot!”
## [1] 0