1.2 Why use ArchR?
There are multiple tools for single-cell ATAC-seq analysis out there so why should you use ArchR? Most importantly, ArchR provides features and enables analyses that other tools do not:
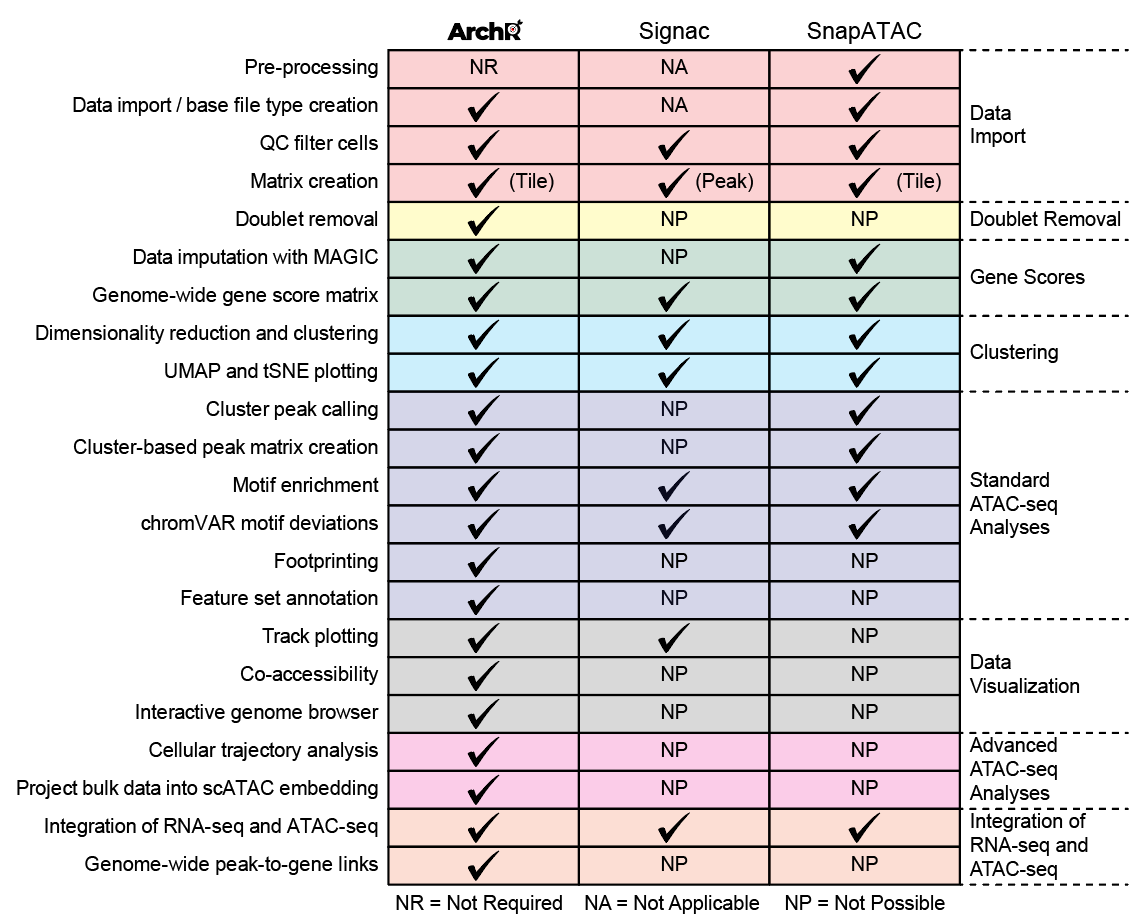
Additionally, ArchR is faster and uses less memory than other available tools due to heavy optimization of the data structures and parallelization methods that form the basis of the ArchR software. When analyzing more than 70,000 cells, some tools require high-performance computing environments, exceeding 128 GB of available memory (OoM = out of memory).
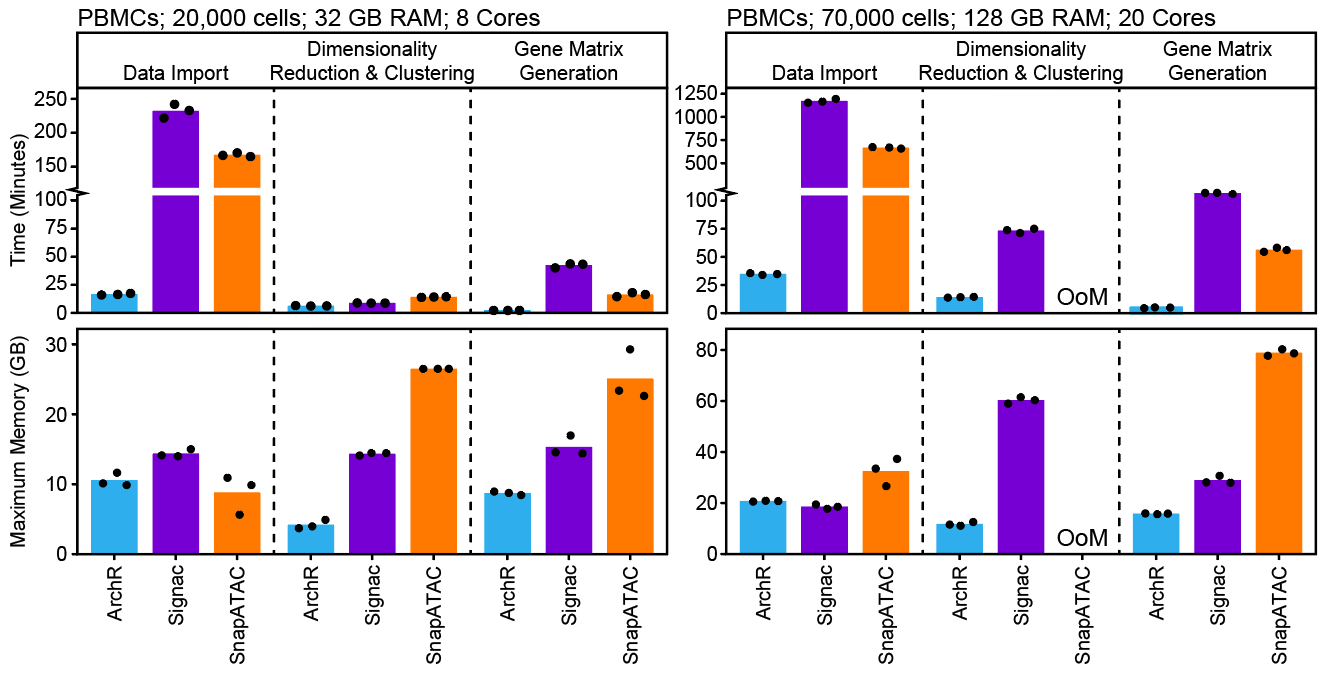
ArchR is designed to be used on a Unix-based laptop. For experiments of moderate size (fewer than 100,000 cells), ArchR is fast enough to perform ad hoc analysis and visualize results in real time, making it possible to interact with the data in a more in-depth and biologically meaningful way. Of course, for higher cell numbers or for users that prefer server-based analysis, ArchR provides fascile export of plots and projects that can be downloaded and used after generation on a server.
Currently, ArchR is not optimized to run on Windows. It should work but parallelization in ArchR has not been enabled for Windows so the performance gains mentioned above will not translate.