4.3 Estimated LSI
For extremely large scATAC-seq datasets, ArchR can estimate the LSI dimensionality reduction with LSI projection. This procedure is similar to the iterative LSI workflow, however the LSI procedure differs. First, a subset of randomly selected “landmark” cells is used for LSI dimensionality reduction. Second, the remaining cells are TF-IDF normalized using the inverse document frequency determined from the landmark cells. Third, these normalized cells are projected into the SVD subspace defined by the landmark cells. This leads to an LSI transformation based on a small set of cells used as landmarks for the projection of the remaining cells. This estimated LSI procedure is efficient with ArchR because, when projecting the new cells into the landmark cells LSI, ArchR iteratively reads in the cells from each sample and LSI projects them without storing them all in memory. This optimization leads to minimal memory usage and further increases the scalability for extremely large datasets. Importantly, the required landmark set size is dependent on the proportion of different cells within the dataset.
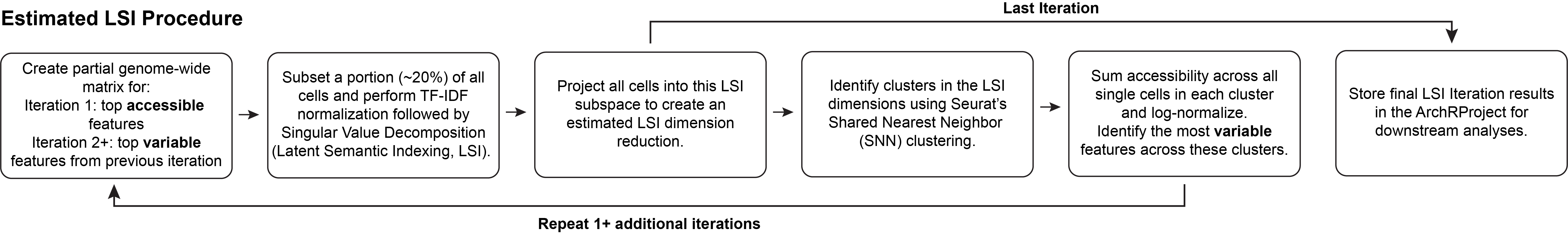
Estimated LSI is accessed in ArchR via the addIterativeLSI() function by setting the sampleCellsFinal and projectCellsPre parameters. samplesCellsFinal designates the size of the landmark cell subset and projectCellsPre tells ArchR to use this landmark cell subset for projection of the remaining cells.