3.3 What is an Arrow file / ArchRProject?
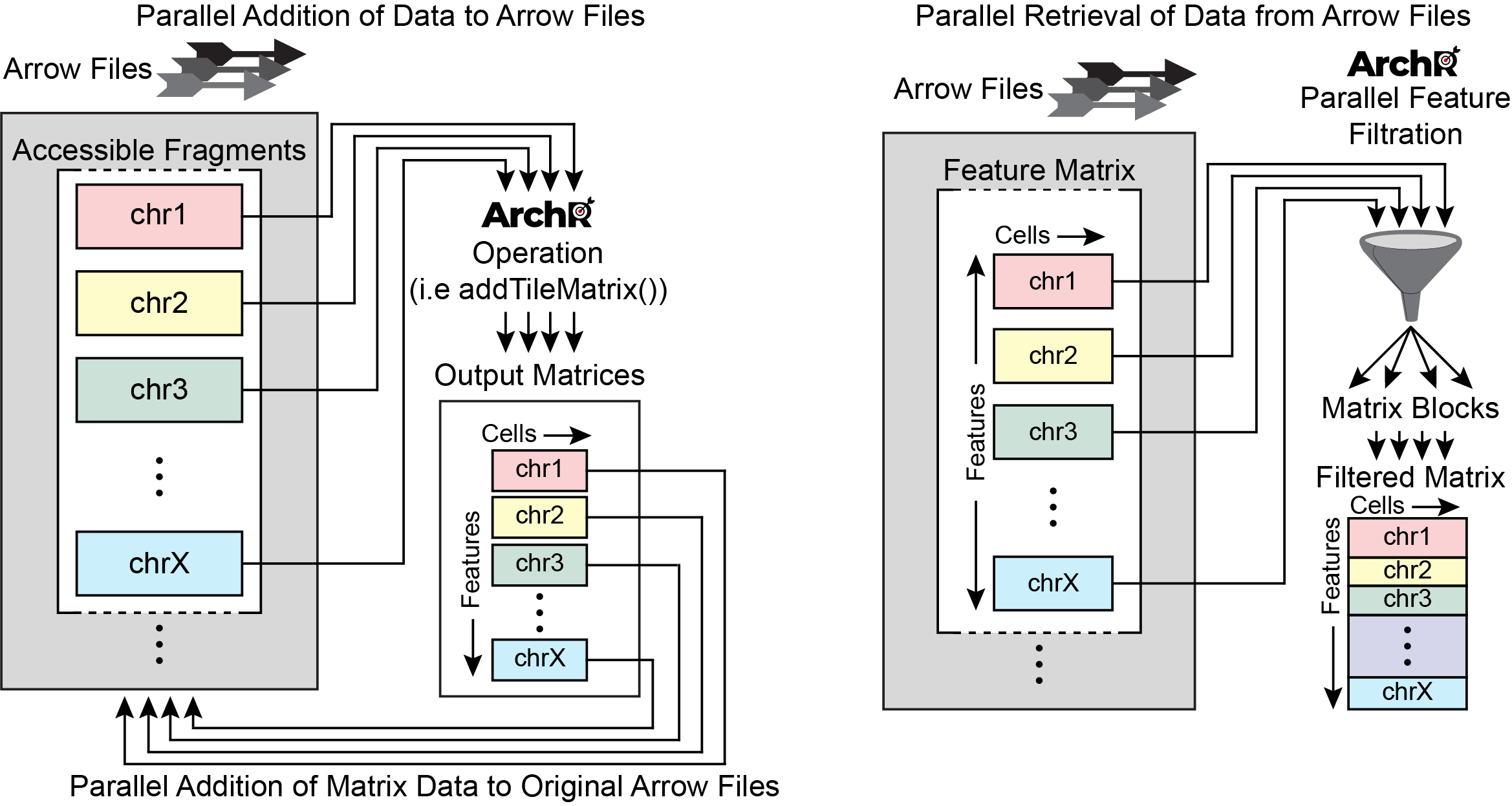
The base unit of an analytical project in ArchR is called an Arrow file. Each Arrow file stores all of the data associated with an individual sample (i.e. metadata, accessible fragments, and data matrices). Here, an “individual sample” would be the most detailed unit of analysis desired (for ex. a single replicate of a particular condition). During creation and as additional analyses are performed, ArchR updates and edits each Arrow file to contain additional layers of information.
It is worth noting that, to ArchR, an Arrow file is actually just a path to an external file stored on disk. More explicitly, an Arrow file is not an R-language object that is stored in memory but rather an HDF5-format file stored on disk. Because of this, we use an ArchRProject object to associate these Arrow files together into a single analytical framework that can be rapidly accessed in R. This ArchRProject object is small in size and is stored in memory.
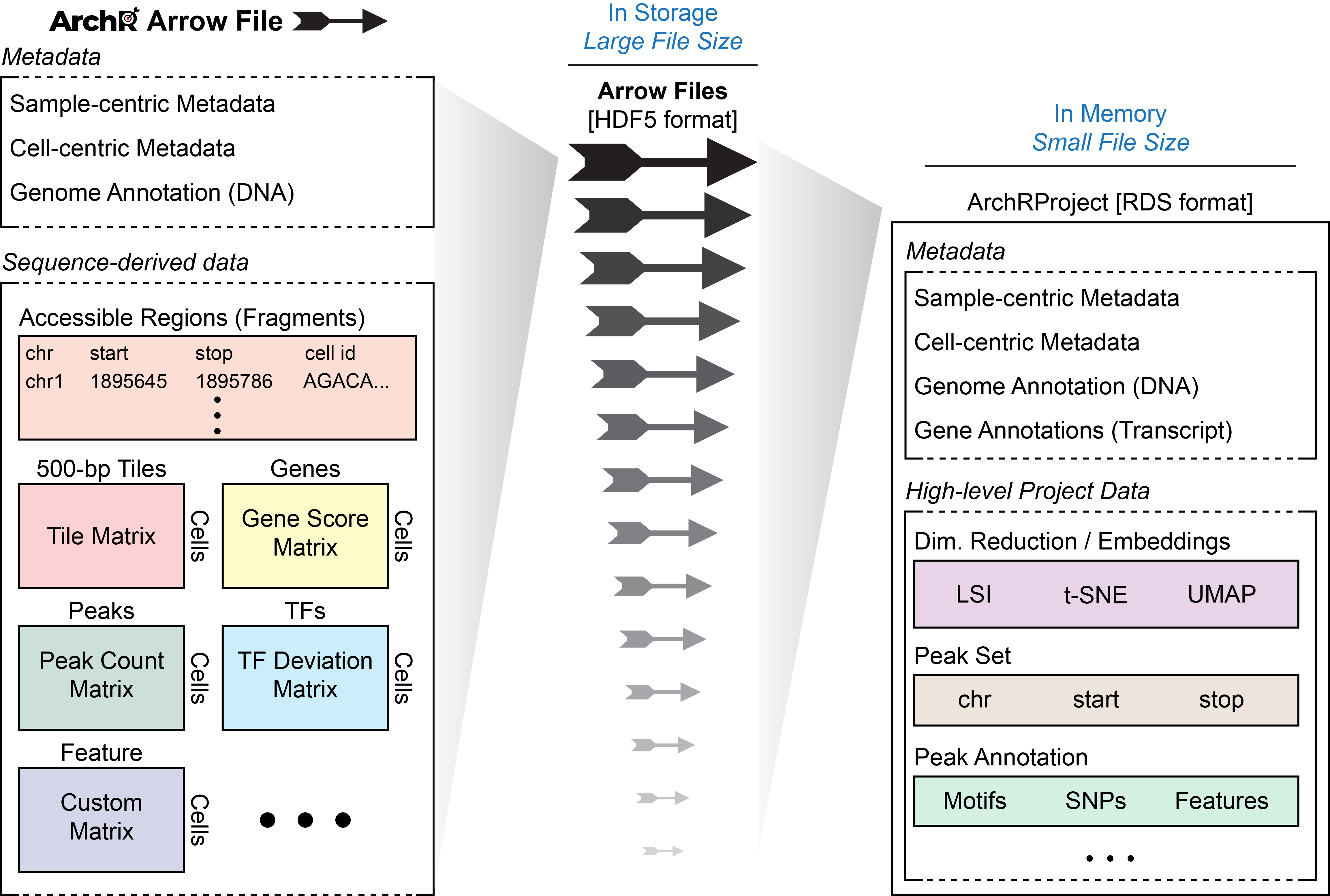
Certain actions can be taken directly on Arrow files while other actions are taken on an ArchRProject which in turn updates each associated Arrow file. Because Arrow files are stored as large HDF5-format files, “get-er” functions in ArchR retrieve data by interacting with the ArchRProject while “add-er” functions either (i) add data directly to Arrow files, (ii) add data directly to an ArchRProject, or (iii) add data to Arrow files by interacting with an ArchRProject.
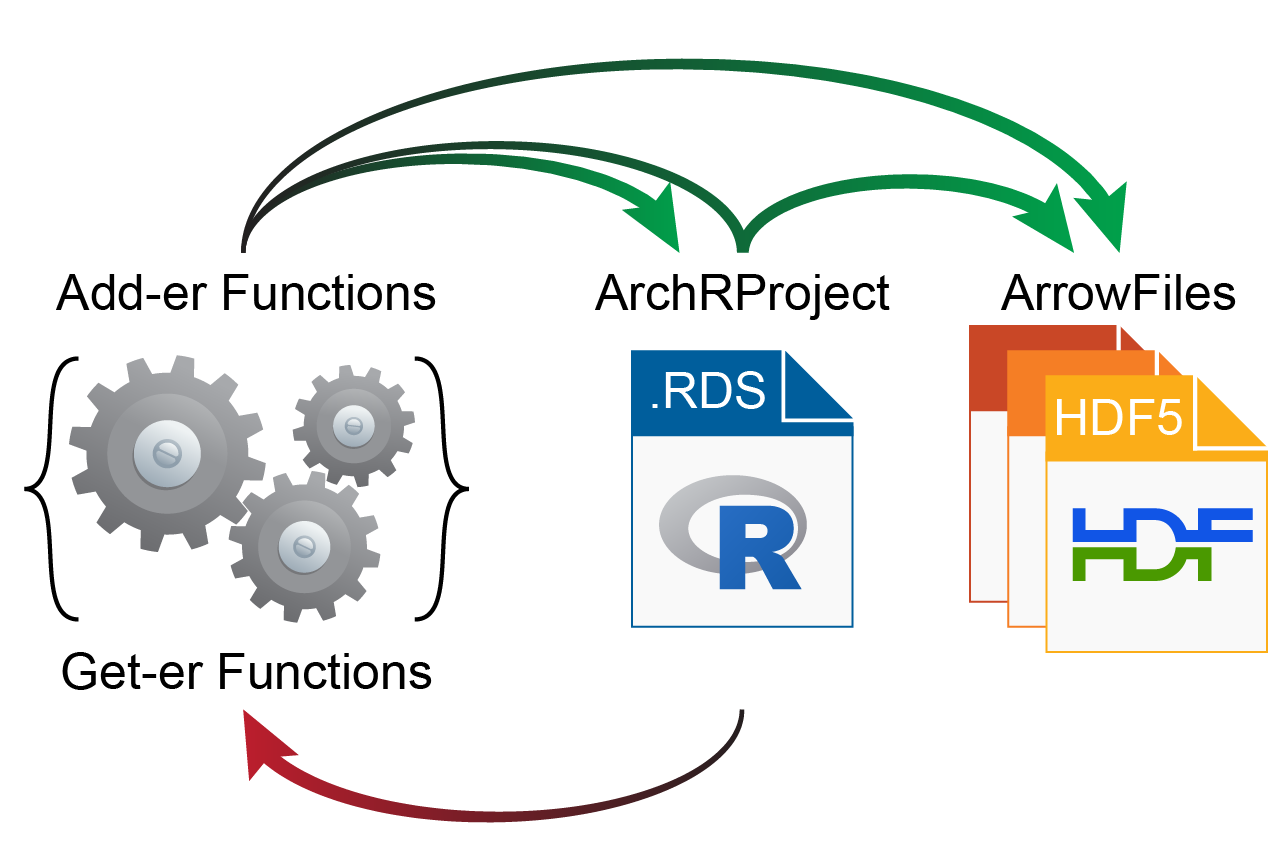
Many of ArchR’s analytical steps can be performed using parallel processing to minimize run-time. This is made possible by the unique data storage employed in Arrow files. Specifically, ATAC-seq fragments are stored per-chromosome and accessed in a chunk-wise fashion.