16.2 Normalization of Footprints for Tn5 Bias
One major challenge with TF footprinting using ATAC-seq data is the insertion sequence bias of the Tn5 transposase which can lead to misclassification of TF footprints. To account for Tn5 insertion bias, ArchR identifies the k-mer (user-defined length, default length 6) sequences surrounding each Tn5 insertion site. To do this analysis, ArchR identifies single-base resolution Tn5 insertion sites for each pseudo-bulk, resizes these 1-bp sites to k-bp windows (-k/2 and + (k/2 - 1) bp from insertion), and then creates a k-mer frequency table using the oligonucleotidefrequency(w=k, simplify.as="collapse") function from the Biostrings package. ArchR then calculates the expected k-mers genome-wide using the same function with the BSgenome-associated genome file. To calculate the insertion bias for a pseudo-bulk footprint, ArchR creates a k-mer frequency matrix that is represented as all possible k-mers across a window +/- N bp (user-defined, default 250 bp) from the motif center. Then, iterating over each motif site, ArchR fills in the positioned k-mers into the k-mer frequency matrix. This is then calculated for each motif position genome-wide. Using the sample’s k-mer frequency table, ArchR can then compute the expected Tn5 insertions by multiplying the k-mer position frequency table by the observed/expected Tn5 k-mer frequency.
All of this happens under the hood within the plotFootprints() function.
16.2.1 Subtracting the Tn5 Bias
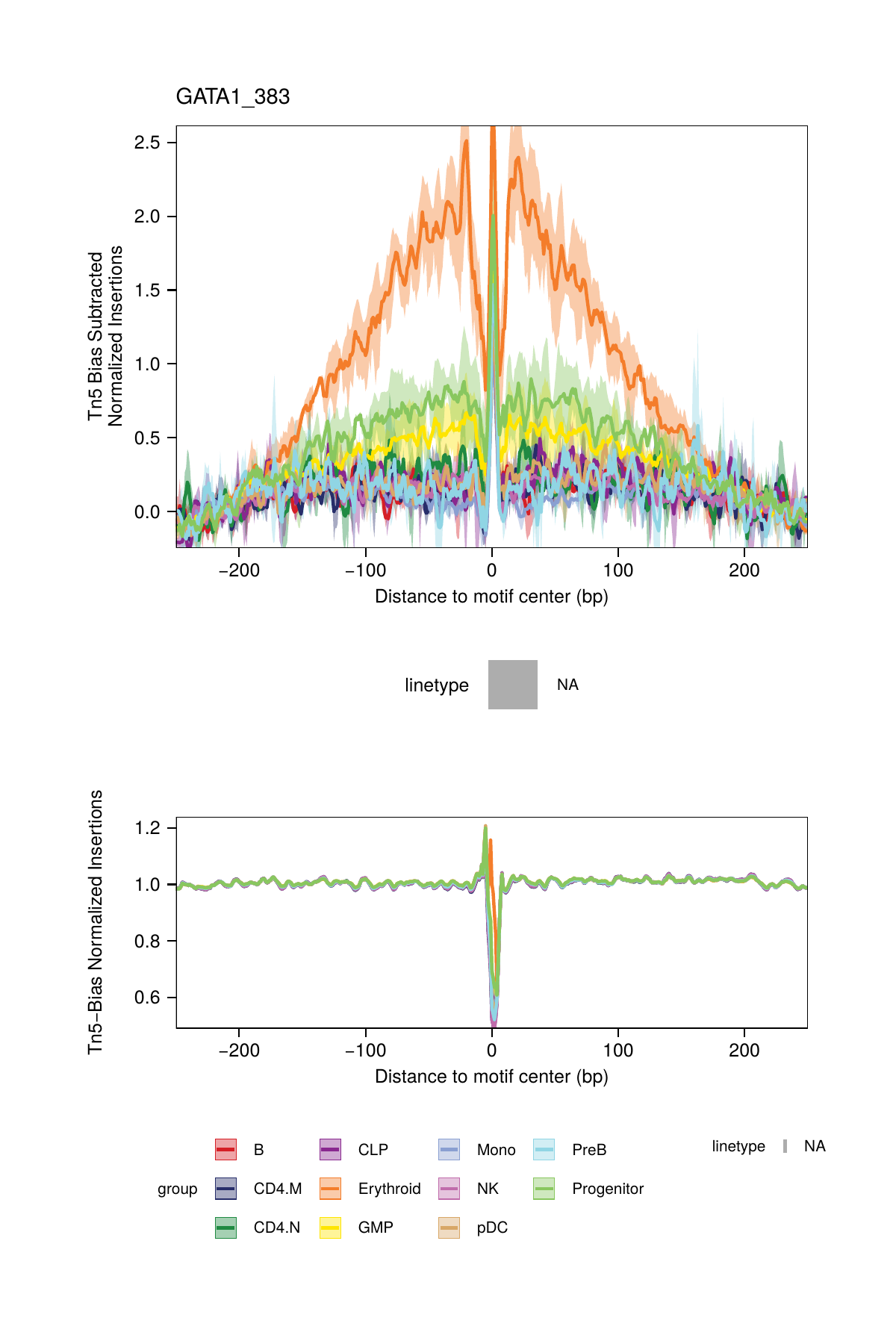
One normalization method subtracts the Tn5 bias from the footprinting signal. This normalization is performed by setting normMethod = "Subtract" when calling plotFootprints().
plotFootprints(
seFoot = seFoot,
ArchRProj = projHeme5,
normMethod = "Subtract",
plotName = "Footprints-Subtract-Bias",
addDOC = FALSE,
smoothWindow = 5
)
## ArchR logging to : ArchRLogs/ArchR-plotFootprints-95a9a2bd7-Date-2025-02-06_Time-02-40-35.7012.log
## If there is an issue, please report to github with logFile!
## 2025-02-06 02:40:35.874202 : Plotting Footprint : GATA1_383 (1 of 6), 0.003 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Subtract
## 2025-02-06 02:40:36.930625 : Plotting Footprint : CEBPA_155 (2 of 6), 0.02 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Subtract
## 2025-02-06 02:40:38.149221 : Plotting Footprint : EBF1_67 (3 of 6), 0.041 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Subtract
## 2025-02-06 02:40:37.414356 : Plotting Footprint : IRF4_632 (4 of 6), 0.029 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Subtract
## 2025-02-06 02:40:38.625704 : Plotting Footprint : TBX21_780 (5 of 6), 0.049 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Subtract
## 2025-02-06 02:40:39.683537 : Plotting Footprint : PAX5_709 (6 of 6), 0.066 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Subtract
## ArchR logging successful to : ArchRLogs/ArchR-plotFootprints-95a9a2bd7-Date-2025-02-06_Time-02-40-35.7012.logBy default, these plots will be saved in the outputDirectory of the ArchRProject. If you requested to plot all motifs and returned this as a ggplot object, this ggplot object would be extremely large. An example of motif footprints from bias-subtracted analyses are shown below.
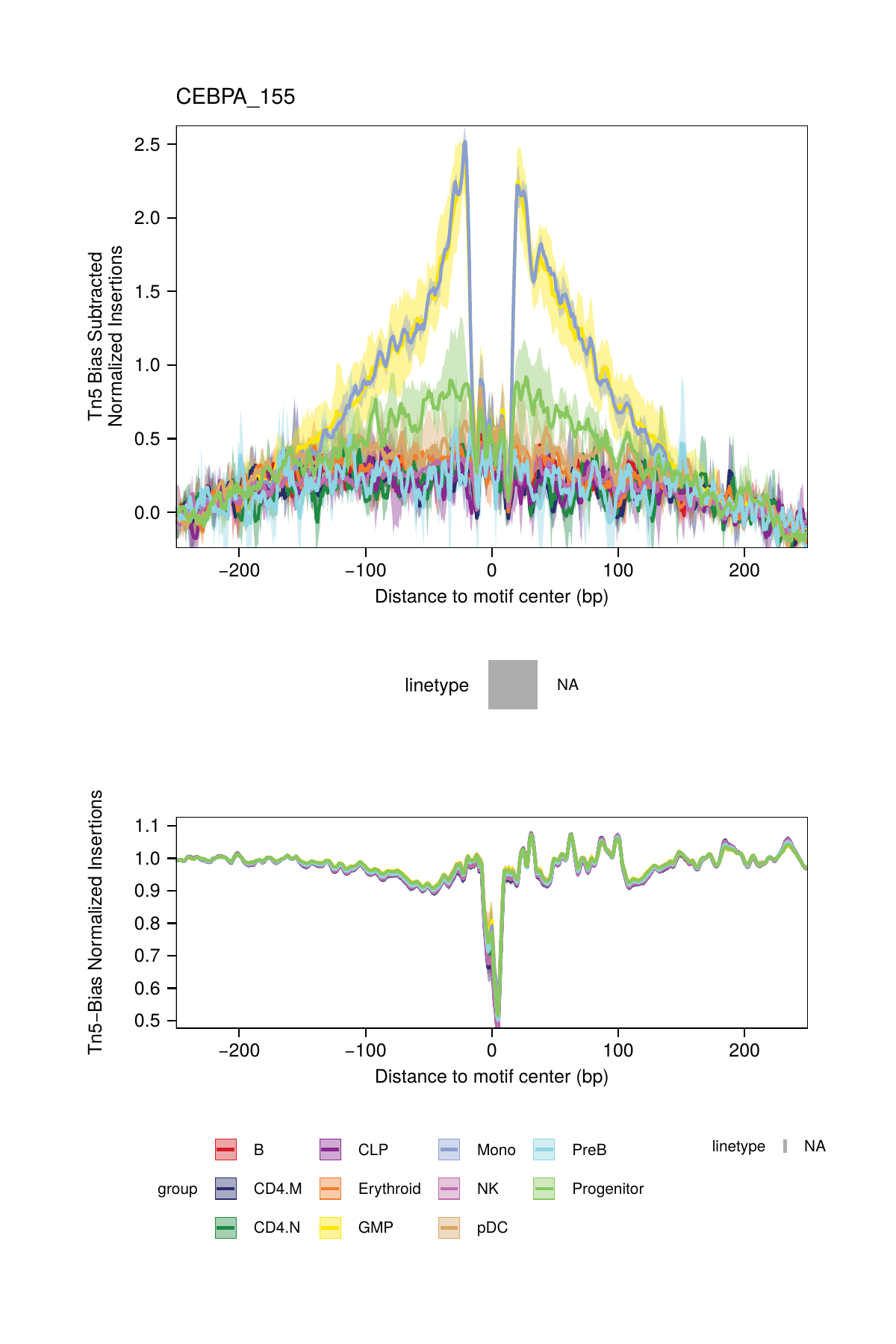
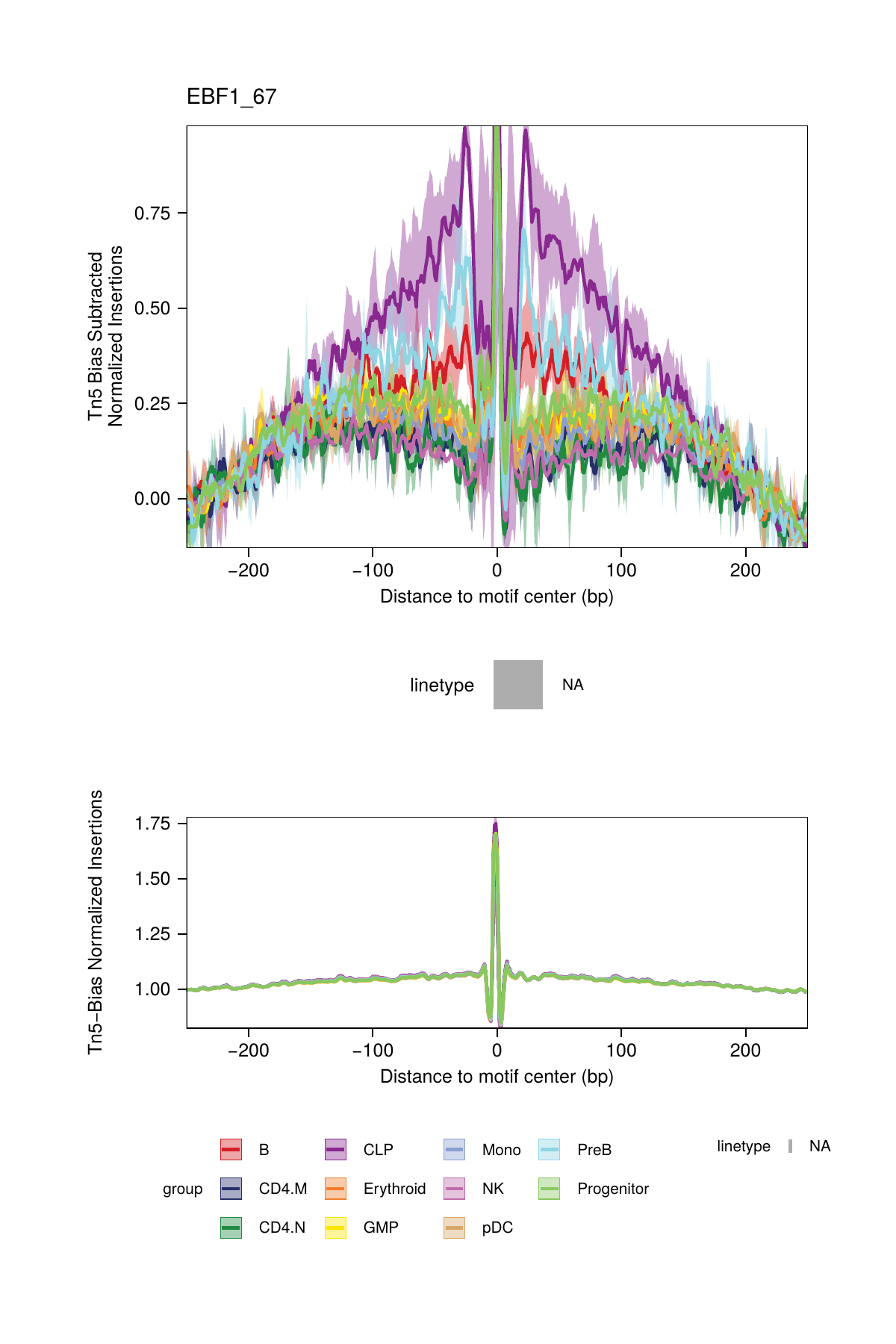
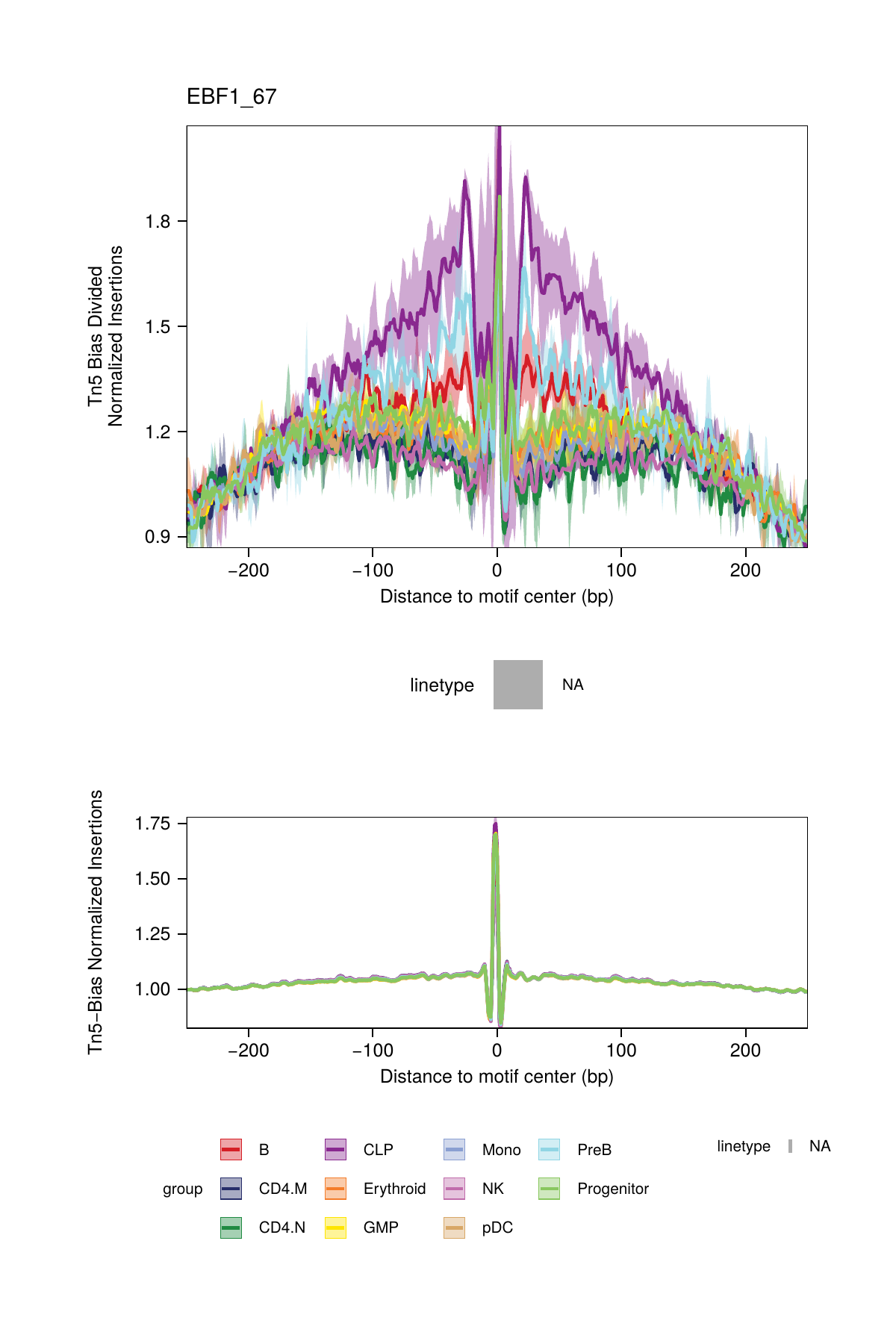
16.2.2 Dividing by the Tn5 Bias
A second strategy for normalization divides the footprinting signal by the Tn5 bias signal. This normalization is performed by setting normMethod = "Divide" when calling plotFootprints().
plotFootprints(
seFoot = seFoot,
ArchRProj = projHeme5,
normMethod = "Divide",
plotName = "Footprints-Divide-Bias",
addDOC = FALSE,
smoothWindow = 5
)
## ArchR logging to : ArchRLogs/ArchR-plotFootprints-93d04dc5-Date-2025-02-06_Time-02-40-44.012551.log
## If there is an issue, please report to github with logFile!
## 2025-02-06 02:40:44.186213 : Plotting Footprint : GATA1_383 (1 of 6), 0.003 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Divide
## 2025-02-06 02:40:45.351897 : Plotting Footprint : CEBPA_155 (2 of 6), 0.022 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Divide
## 2025-02-06 02:40:46.601008 : Plotting Footprint : EBF1_67 (3 of 6), 0.043 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Divide
## 2025-02-06 02:40:47.846978 : Plotting Footprint : IRF4_632 (4 of 6), 0.064 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Divide
## 2025-02-06 02:40:49.30056 : Plotting Footprint : TBX21_780 (5 of 6), 0.088 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Divide
## 2025-02-06 02:40:50.731304 : Plotting Footprint : PAX5_709 (6 of 6), 0.112 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = Divide
## ArchR logging successful to : ArchRLogs/ArchR-plotFootprints-93d04dc5-Date-2025-02-06_Time-02-40-44.012551.logAn example of motif footprints from bias-divided analyses are shown below.
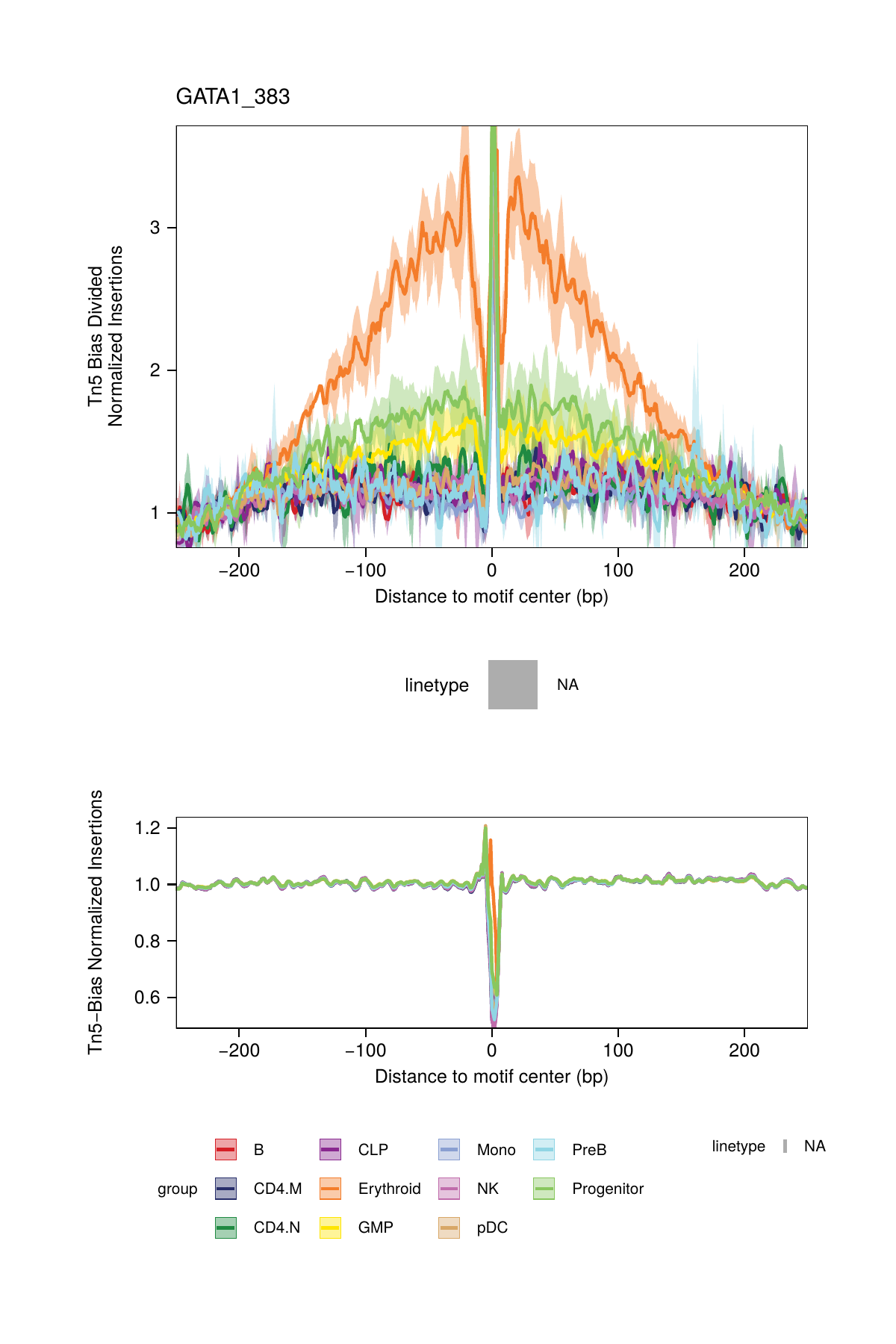
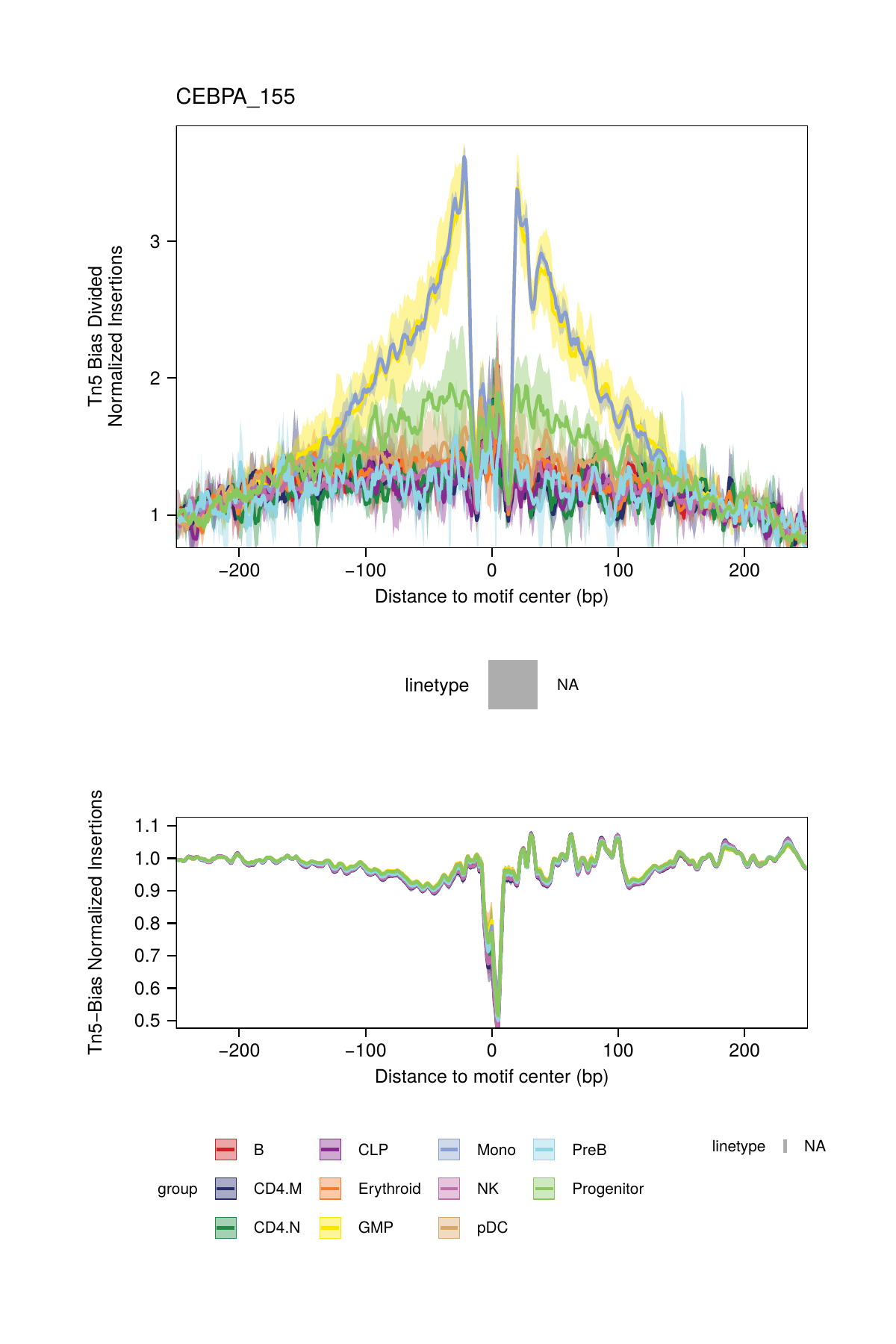
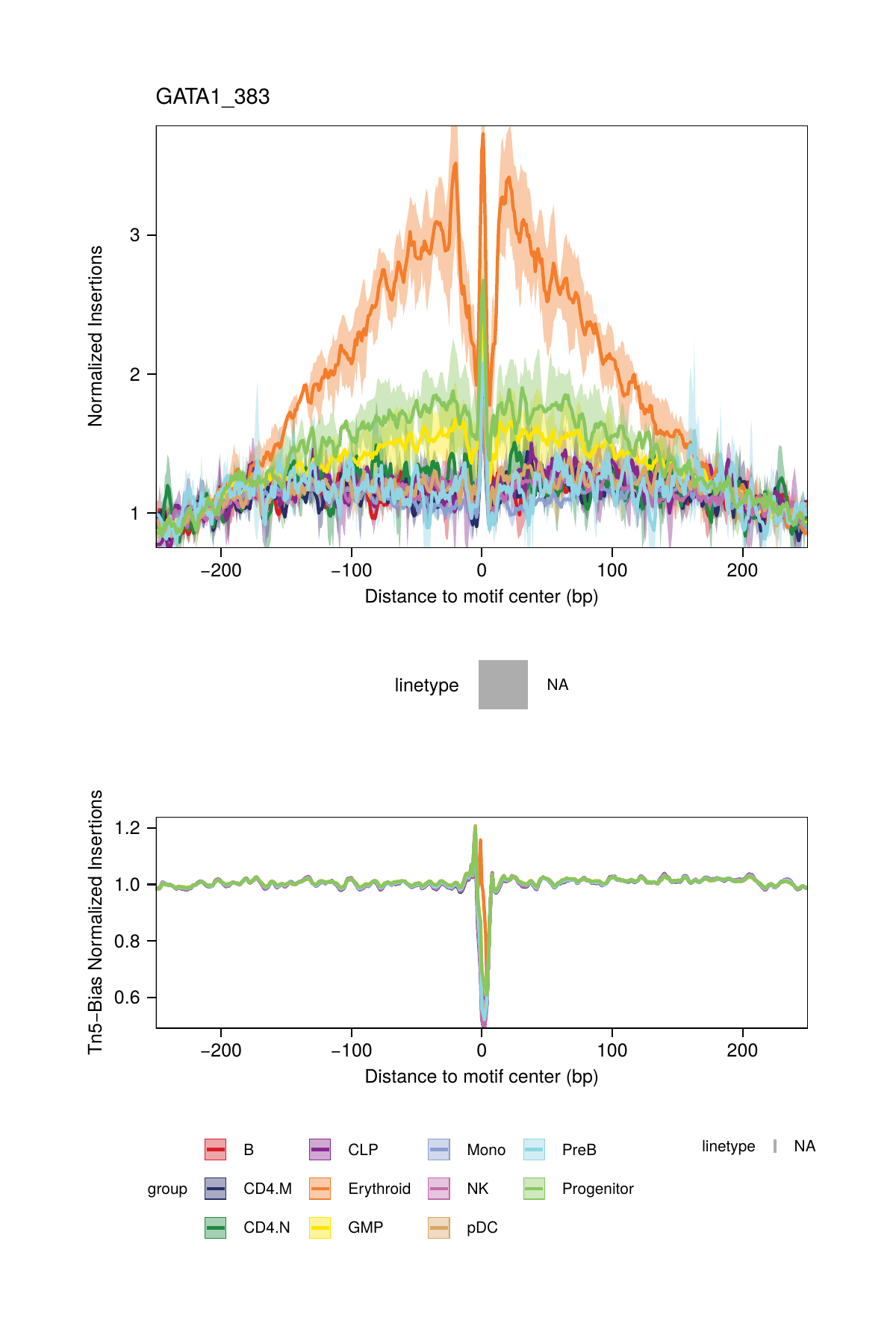
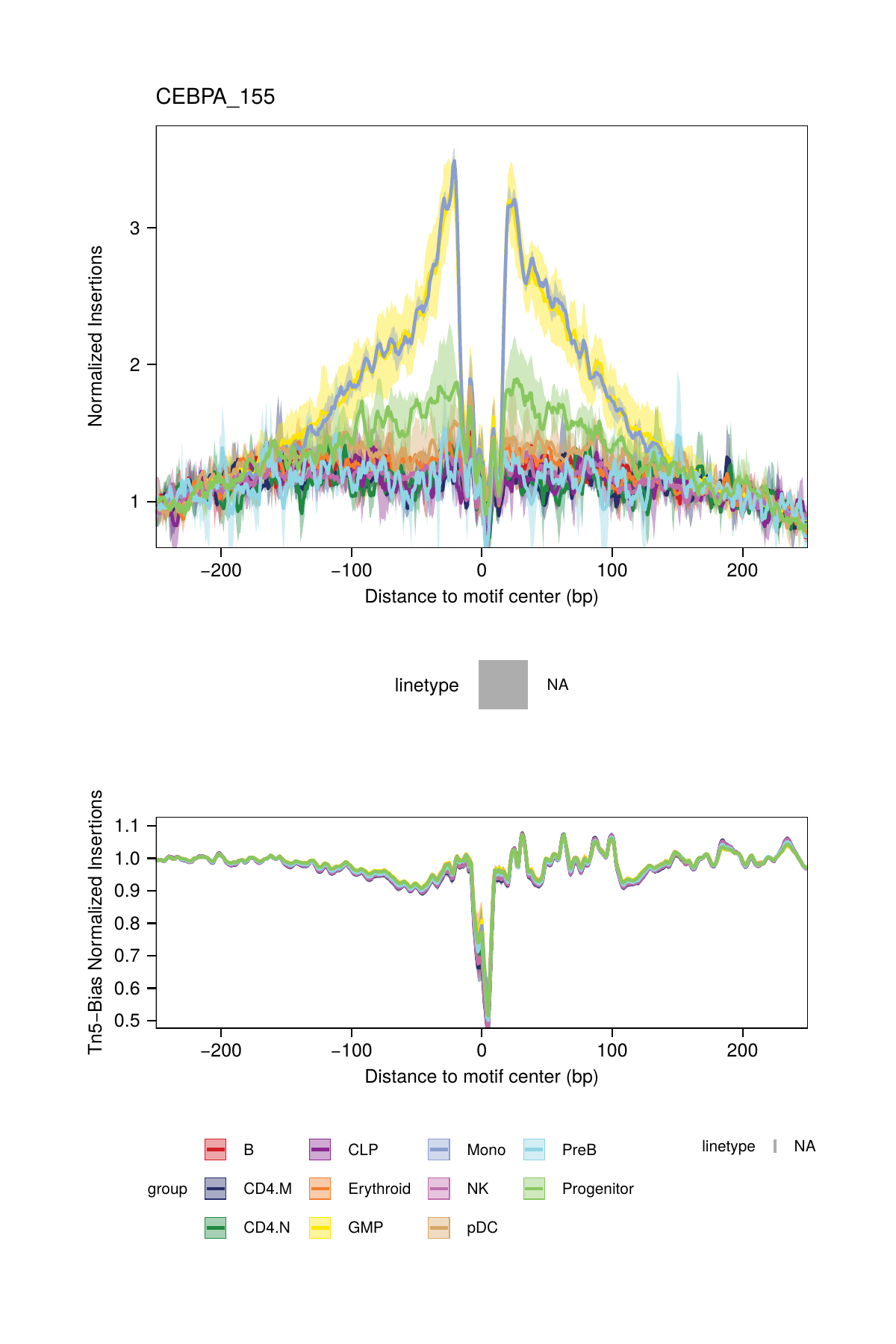
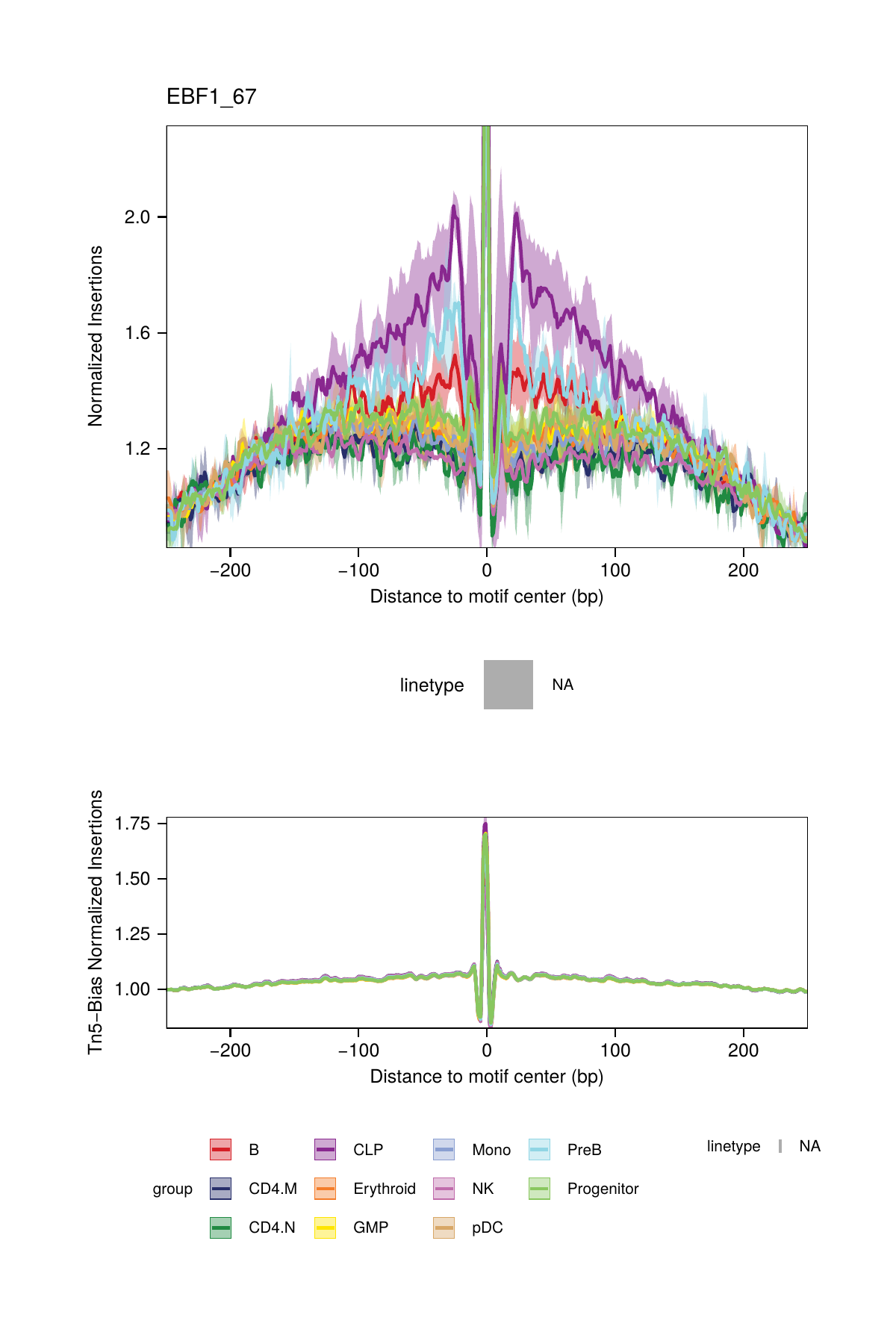
16.2.3 Footprinting Without Normalization for Tn5 Bias
While we highly recommend normalizing footprints for Tn5 sequence insertion bias, it is possible to perform footprinting without normalization by setting normMethod = "None" in the plotFootprints() function.
plotFootprints(
seFoot = seFoot,
ArchRProj = projHeme5,
normMethod = "None",
plotName = "Footprints-No-Normalization",
addDOC = FALSE,
smoothWindow = 5
)
## ArchR logging to : ArchRLogs/ArchR-plotFootprints-94f0ac140-Date-2025-02-06_Time-02-40-54.965769.log
## If there is an issue, please report to github with logFile!
## 2025-02-06 02:40:55.146243 : Plotting Footprint : GATA1_383 (1 of 6), 0.003 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = None
## 2025-02-06 02:40:56.203805 : Plotting Footprint : CEBPA_155 (2 of 6), 0.021 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = None
## 2025-02-06 02:40:57.43399 : Plotting Footprint : EBF1_67 (3 of 6), 0.041 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = None
## 2025-02-06 02:40:58.622308 : Plotting Footprint : IRF4_632 (4 of 6), 0.061 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = None
## 2025-02-06 02:40:59.826235 : Plotting Footprint : TBX21_780 (5 of 6), 0.081 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = None
## 2025-02-06 02:41:00.904494 : Plotting Footprint : PAX5_709 (6 of 6), 0.099 mins elapsed.
## Applying smoothing window to footprint
## Normalizing by flanking regions
## NormMethod = None
## ArchR logging successful to : ArchRLogs/ArchR-plotFootprints-94f0ac140-Date-2025-02-06_Time-02-40-54.965769.logAn example of motif footprints without normalization are shown below.